Copy Number Variation in the Human Genome and its Role in Human Evolution
NOTE: Apologies, the images I have uploaded appear to be a little difficult to decipher on a black background. In order to make good use of them you may need to save them and open them in an image viewer. – Confusedious
Introduction
Following the completion of the human genome project in 2003 (Collins, Morgan, & Patrinos, 2003), and the technological advances in gene sequencing and mapping this project enabled (Watson, 2004), it became apparent that the human genome was host to great inter-individual and inter-population variation. While much of the work on human genetic diversity has focused upon variations at the nucleotide level, the so-named point mutations or single nucleotide polymorphisms (SNPs) (Freeman et al., 2006), researchers have also come to realise that polymorphisms of a larger scale are both relatively abundant and of importance in terms of phenotypic expression (Fu, Zhang, Wang, Gu, & Jin, 2010), flagging them as being of potential consequence to studies of human evolution (Zhang, Gu, Hurles, & Lupski, 2009). Prime among these larger scale polymorphisms are those known as copy number variations (CNVs), an umbrella term used to refer to the duplication or deletion of regions of DNA of variable length (from a few hundred base pairs to millions of base pairs), often with consequences for the products of the genes encoded within, and thus, observable impacts upon phenotype that can be considered to alter fitness (Freeman, et al., 2006; Zhang, et al., 2009).
While much of the work on CNVs to date has focused upon simply identifying sites of variation in the human genome and comparing these loci in terms of frequency and positioning on chromosomes between individuals of differing geographic ancestry for ‘genealogical’ purposes (Chen et al., 2011; Li et al., 2009; Redon et al., 2006), or for the investigation of pathophysiologies (Horev et al., 2011; Lee & Lupski, 2006; Mills et al., 2011), this essay shall focus upon the evolutionary implications of CNVs in the human genome from both an inter and intra specific position. In order to better place CNVs in an evolutionary context, the first section of this essay shall focus upon how these variations occur and produce phenotypic changes that can be of evolutionary consequence. Following this, an analysis of CNVs between Homo sapiens and our nearest extant relative, Pan troglodytes, shall be assessed for what they can tell us about the broader evolutionary history of hominins. Moving to a focus on relatively recent human evolutionary history, the role of CNVs in adaptation to the unique problems faced by post-Neolithic revolution populations shall be explored, namely the problems of dietary change and pathogen burden as a result of higher density living. In closing, the principal difficulties in the interpretation of CNVs in the context of human evolution shall be highlighted and predictions about the findings of future studies shall be made.
How Copy Number Variations Arise and Produce Phenotypic Change
Prior to the completion of the human genome project, CNVs were known to exist but were assumed to be relatively rare and, as such, relatively unimportant (Freeman, et al., 2006). Recent estimates, however, place the percentage of the human genome that is subject to these variations at between ~12% (Hastings, Lupski, Rosenberg, & Ira, 2009) and an impressive ~30% (Zhang, et al., 2009), a significantly higher proportion than that of <1% in SNPs (Zhang, et al., 2009). Moreover, the rate of de novo CNVs is estimated to be between 100 and 10,000 times greater than that for SNPs depending on the region of the genome being examined (Fu, et al., 2010). Given that SNPs seem rare when compared to CNVs, perhaps due to the high likelihood of them causing deleterious coding errors (nonsense code, frameshift etc.) (Zhang, et al., 2009) and thus being eliminated by purifying selection, the mechanism behind the seemingly more ‘stable’ CNVs is of exceptional interest to those wishing to understand evolution at the genetic level.
One does, however, need to exercise a certain degree of caution in approaching CNVs as potentially positively selected mutations. The fact that, at the surface level, CNVs seem to be more abundant and stable should not be taken to mean that they are inherently more evolutionarily advantageous than SNPs, in fact many CNVs are thought to be neutral or somewhat deleterious (linked to disease states), it may just be that purifying selection acts with greater force on SNPs due to their capacity to disrupt normal gene expression at a more fundamental level, when compared to the inherent structural stability of a length of otherwise functional DNA that has simply been duplicated wholesale (Hastings, et al., 2009). Wholesale duplications (or deletions) of this kind have been argued to be the consequence of two related processes, homologous recombination (HR) and non-homologous recombination (NHR) (Hastings, et al., 2009).
HR and NHR are processes that act to repair double strand breakages in DNA through, aligning and reattaching lengths of DNA that ‘match’ those leading up to the breakage in the case of HR, or through attaching lengths of DNA that do not necessarily match in the case of NHR (Hastings, et al., 2009; Lieber, Ma, Pannicke, & Schwarz, 2003). In the case of HR, due to all human genomes containing regions of duplication called segmental duplications (SDs) (Lacroix, Oparina, & Mashkova, 2003) (these differ from CNVs in that these regions are generally not polymorphic), breakages in these areas can result in sequences from another section of any given chromosome, with near parity, being attached to this site, resulting in duplication (Hastings, et al., 2009) (Figure 1). This results in a net increase in copy number of any genes present. Alternatively, this same mechanism can result in a net loss (deletion) should intermediate genetic material be sheared when a near homologous strand from elsewhere in the genome, that lacks this intermediate material, is used to repair a break. As well as occurring in the repair of broken double stranded DNA, similar unequal exchanges can occur during unequal meiotic cross-over events as a function of HR, also potentially resulting in CNVs (Hastings, et al., 2009) (Figure 1). NHR functions similarly in that it rejoins broken double stranded DNA, with the exception that it does not require the same degree of homology between the two strands to be joined and can thus be considered a more efficient process despite the higher likelihood of error (Guirouilh-Barbat et al., 2004; Hastings, et al., 2009; Lieber, et al., 2003). This process is more common in mammals than HR (Guirouilh-Barbat, et al., 2004) and, as such, can be seen to be one of the main forces behind CNVs in the human genome.
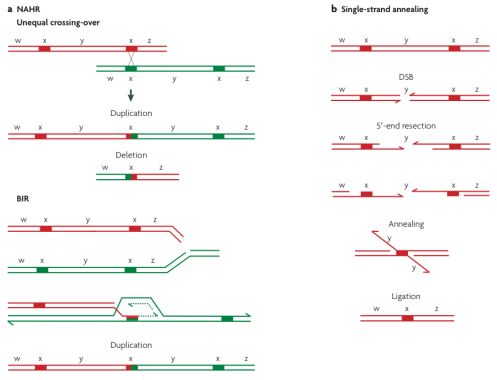
Figure 1. How deletions and duplications can occur during unequal meiotic cross-over (a) and how deletions can occur as a result of repair by homologous recombination of broken double stranded DNA (b). Adapted from Hastings et al. (2009).
As mentioned, these changes in copy number are capable of producing phenotypic differences in individuals. These phenotypic impacts are principally produced through ‘dosage’ effects, or put simply, an increase or decrease in the volume of the gene product (structural protein, hormone, enzyme etc.) being produced as a consequence of changes in activity in the production or regulation of this product due to the duplication or deletion of related genes (Perry, 2008; Zhang, et al., 2009). It must be noted, however, that these effects are not linearly related to the copy number of the gene in question due to the fact that gene expression is a complex process that relies on regulatory sequences to influence the degree to which any given gene is expressed, if it is expressed at all. For example, CNV in the OPN1MW gene, associated with colour vision, is relatively common but it is only the copy nearest the intact regulatory sequence that is of consequence (Perry, 2008). Should an individual have a dysfunctional copy of this gene closest to the regulatory sequence, any additional copies of the functional gene elsewhere in the genome will not be expressed with the result that the individual will be colour-blind (Perry, 2008). This being said, there are numerous genes that are subject to CNVs that have known and observable dosage dependent effects, many of which will be discussed below.
Any genetic polymorphism that results in phenotypic characters that offer benefits or hindrances, in the survival stakes, to those carrying them can be said to influence evolutionary fitness. It stands to reason that should any given CNV be of advantage to fitness, it should increase in frequency among the population on whom it confers this advantage. Beginning with a comparison of the human genome to that of our closest extant relative, the chimpanzee (P. troglodytes), evidence shall be sought that CNVs have, indeed, played a crucial role in the divergence of these two species from a common ancestor some ~6 million years ago.
Copy Number Variation and the Divergent Evolution of Pan and Homo
Recent comparisons between the human and chimpanzee genomes have yielded a wealth of information about the respective evolutionary pasts of these two species. In a comparison of SDs present in the human and chimpanzee genomes, Cheng et al. (2005) have suggested that only 33% of these non-polymorphic duplications were not present in the chimpanzee genome, suggesting a high degree of similarity in terms of chromosomal geography when it comes to retained regions of duplication. This is of interest as it does suggest that the common ancestor of both of these species shared many of these SDs, and with regions of SD being ‘hotbeds’ for de novo CNV type mutations (SDs themselves are simply CNVs that have become relatively fixed) (Fu, et al., 2010; Mills, et al., 2011; Perry et al., 2008), this provides researchers with an understanding of where to focus when searching for CNVs.
Interpreting CNV similarities or differences between two species thought to have diverged so long ago, however, must be done with care. Perry et al. (2008) have convincingly argued that due to the relative instability of the regions that CNVs tend to inhabit and the rapid de novo mutation rate of CNVs, that any such observable similarities in CNVs (as opposed to the more consistent SDs) between these two genomes are likely the result of independent and relatively recent convergent evolutionary events rather than ancestral genomic traits maintained for ~6 million years. However, differences and similarities between patterns of genes enhanced or blocked through CNVs still can provide information about the differing evolutionary pressures experienced by these two species and offer insights into how, in terms of biological machinery, contemporary phenotypes were produced. Some of the CNVs of interest that have featured in recent work shall be discussed below.
In a study that utilised genomic comparisons across ten primate species, including humans and chimpanzees, Dumas et al. (2007) identified numerous CNVs in the human genome that seemed to correspond well with our palaeontolgoical understanding of hominin evolution. The increase in brain size and complexity in the hominin lineage, particularly within the last two million years, is a well accepted part of contemporary human evolutionary models (Navarrete, van Schaik, & Isler, 2011). In their study, Dumas et al. (2007) found a marked increase in the duplications of DNA containing the gene DUFF1220 when compared to other primate lineages. DUFF1220 is a gene thought to be related to higher cognitive processes based on the fact that damage to this gene results in mental retardation both in humans and experimental mice (Dumas, et al., 2007), suggesting that duplications of this gene in our evolutionary past may have contributed to the development of more sophisticated cognitive capabilities. Likewise, human specific duplications of NEK2 and ANAPC1 were also observed, these two genes being implicated in mitotic division and thought to be possible affecters of neocortex expansion in the hominin lineage (Dumas, et al., 2007). Furthermore, numerous models of human evolution have emphasised the role of endurance running in human specific traits such as sweating and fat metabolism, aspects of our evolutionary past that Dunbar et al. (2007) have argued may have been facilitated by copy number increases of AQP7. This gene is thought to be implicated in water and glycerol transport across cell membranes, and as such is of potential importance in both the evolution of more efficient extraction of energy from stored body fat and sweating as a response to overheating (Dumas, et al., 2007). It must be noted here, however, that the importance of ‘endurance’ or ‘persistence’ hypotheses of human evolution are not universally agreed upon, so other explanations for the increase in copy number of AQP7 may be possible.
In addition to the duplications identified by Dumas et al. (2007), a more recent genomic comparison between humans and chimpanzees by McLean et al. (2011) has identified 510 human lineage specific deletions. While many of these genetic losses fall within non-coding regions, numerous were associated with the deletion of regulatory sequences that are otherwise highly conserved among primates (McLean, et al., 2011). Two of these regulatory deletions were of particular interest, the first affecting expression of penile spines (associated with the androgen receptor gene) and the second relating to the expression of GADD45G, a tumor suppression gene, in parts of the brain thought to be related to the generation of cells responsible for neocortex expansion (McLean, et al., 2011). The absence of penile spines in humans can be argued to be related to changes in sexual behaviour in hominins, with one replier to McLean et al. (2011) suggesting that the reduction in sexual sensitivity this would cause could be associated with increased coital duration, generating greater bonding between the sexes in a lineage that was becoming increasingly monogamous (van Driel, 2011). As previously mentioned, the expansion of the brain in the human lineage is an area of importance, and the deletion of regulators that restrict brain size, in particular that of the neocortex, are of obvious significance when attempting to piece together the puzzle of brain size increase in the hominin line.
Taken together, it is clear that work within the last decade on comparing genetic duplications and deletions between the human and chimpanzee genomes has yielded significant insight into how observable phenotypic differences between these species have come to exist. While explanations of how these genetic changes came to be selected are likely to never be entirely agreed upon, it would appear that these changes do fit with several existing palaeontological hypotheses of how the Homo line evolved, particularly where brain expansion, endurance running and sexual behaviours are concerned.
Copy Number Variations in the Human Genome and Recent Human Evolution
Moving in focus from the divergence of our species from that of our nearest neighbour millions of years ago and the subsequent development of the hominin line, CNVs in the human genome can also tell us much about selective pressures in our recent evolutionary history. A particular application of this type of investigation is in considering how the adoption of agriculture and, consequently, higher density urban living following the Neolithic revolution ~10,000 years ago has produced selective pressures that have shaped the genomes of those populations involved (Richerson, Boyd, & Henrich, 2010). Here we shall focus on how dietary changes following the Neolithic revolution have generated a particularly well studied CNV and how the increased infectious disease burden of urban living has also generated a beneficial genetic duplication.
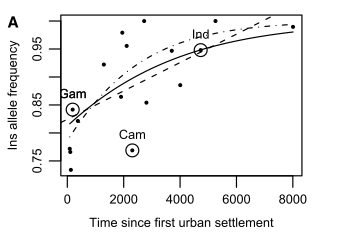
A particular characteristic of the diet of agricultural societies is their reliance on high starch grains such as wheat, millet, rice and barley (Diamond, 1991). Working from this fact, Perry et al. (2007) made an investigation of the correlation between CNVs in AMY1, the dose sensitive gene responsible for the production of salivary amylase, an enzyme utilised in the breakdown of starch, and dietary intake of starch across various populations including Europeans, Japanese, Hadza, Mbuti, Biaka, Datog and Yakut. It was found that the three populations with the greatest number of copies of the AMY1 gene were two agricultural societies, the Europeans and Japanese, and the Hadza, that relied less upon agriculture but had a high percentage of tubers and other starch rich foods in their diets (Novembre, Pritchard, & Coop, 2007; Perry, et al., 2007) (Figure 2). Interestingly, Mandel et al. (2010) have also found that increased copy numbers of AMY1 favourably affect the perception of the texture of starchy foods, increasing their desirability as a consistent part of the diet. These findings strongly suggest that relatively recent diet changes have produced sufficient selective pressures as to generate the accumulation of gene duplications that facilitate both the digestion and textural perception of important subsistence diet items.
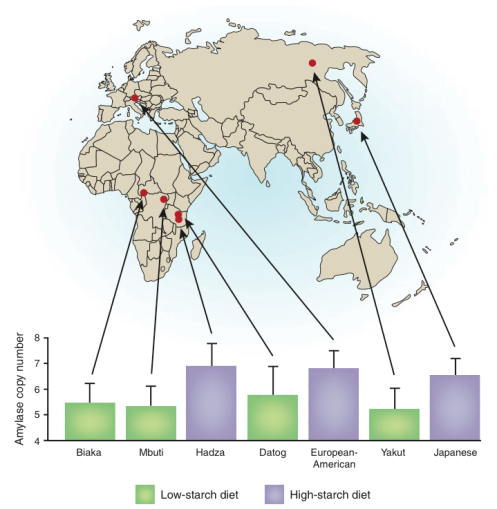
Figure 2. The correlation between salivary amylase copy number and low or high starch diets in the study populations utilised in Perry et al. (2007). Novembre et al. (2007).
The development of higher density living arrangements (towns and eventually cities) as a consequence of increasingly efficient agricultural practices, in parts of the world such as the Fertile Crescent, North Africa, Europe, India and East Asia, brought with it an increase in the infectious disease burden (Diamond, 1991). Based on recent evidence to be discussed here, this can be argued to have generated sufficient selective pressure as to favour those individuals with CNVs that offered some resistance to these diseases. Recent research by Hardwick et al. (2011) into the geographic distribution copy numbers of a high expressing version of a gene associated with the antimicrobial properties of epithelial tissue (beta-defensin, DEFB103), found that high copy numbers of this gene were concentrated in East Asian populations. Given the historically high burden of influenza and other zoonotic viral infections of the epithelium in this area, in addition to the relatively long history of high density population centres, high copy numbers of this gene were argued to have accumulated as a consequence of their conferral of greater resistance to these infections (Hardwick, et al., 2011). Given other recent research into a non-CNV type polymorphism, thought to confer greater resistance to tuberculosis (Barnes, Duda, Pybus, & Thomas, 2011), that has shown a strong frequency correlation with populations with a long history of urban living, the explanation offered by Hardwick et al. (2011) of the high copy numbers of the high expressing DEFB103 variant, seen in East Asia, seem quite plausible.
While only two CNVs have been offered here as having strong correlations with relatively recent human lifestyle changes, namely niche construction in the form of agriculture and urbanisation, weight can be added to these examples by referring to the more abundant data on SNPs thought to be associated with post-Neolithic revolution changes in selective pressures. Studies by Wang et al. (2006) and Hawks et al. (2007) have identified hundreds of genes on which, based on linkage disequilibrium, strong selection seems to have been operating within the last ~40,000 years, with Hawks et al. (2007) suggesting that many of these are likely to have occurred within the last ~10,000 years as a consequence of post-Neolithic revolution lifestyle changes and population expansions. Based on this, it seems highly probable that many more CNVs relating to adaptations to a ‘modernising world’ are likely to be identified in the human genome of those populations impacted by these changes.
Conclusion
In summary, the inter and intra species study of copy number variations in the human genome can tell us much about the genetic machinery that underlies the observable phenotypic differences between modern humans and our nearest extant relative the chimpanzee, as well as those between modern human geographical populations. Additionally, evidence of positive selection for certain gene configurations, be they duplications or deletions, in the past can offer some insight into the selective pressures experienced by ancestral hominin populations. While it is estimated here that future studies are likely to identify further examples of CNVs useful for the purposes of understanding human evolution, both recently and in the distant past, such data must always be interpreted carefully due to the fact that the de novo mutation rate of CNVs is relatively high, reducing the likelihood that any observably frequent copy number variant is indeed ancestral. Additionally, while this essay has largely omitted the technical details of how CNVs are identified in the human genome, the focus of much contemporary research on the identification of SNPs does relatively little to aid the advancement of techniques useful for more efficient CNV identification. Should the study of CNVs gain anything like the momentum experienced in the study of SNPs, it is predicted here that the subsequent advances in detection techniques should yield a marked increase in the discovery of the kinds of CNVs described here, greatly enhancing our understanding of how this particular genetic mutation process has contributed to human evolution.
References
Barnes, I., Duda, A., Pybus, O. G., & Thomas, M. G. (2011). Ancient Urbanization Predicts Genetic Resistance to Tuberculosis. Evolution, 65(3), 842-848.
Chen, W., Hayward, C., Wright, A. F., Hicks, A. A., Vitart, V., Knott, S., et al. (2011). Copy Number Variation across European Populations. PLoS ONE, 6(8), e23087.
Cheng, Z., Ventura, M., She, X., Khaitovich, P., Graves, T., Osoegawa, K., et al. (2005). A genome-wide comparison of recent chimpanzee and human segmental duplications. [10.1038/nature04000]. Nature, 437(7055), 88-93.
Collins, F. S., Morgan, M., & Patrinos, A. (2003). The Human Genome Project: Lessons from Large-Scale Biology. Science, 300(5617), 286-290.
Diamond, J. (1991). Guns, Germs and Steel. London: Jonathan Cape.
Dumas, L., Kim, Y. H., Karimpour-Fard, A., Cox, M., Hopkins, J., Pollack, J. R., et al. (2007). Gene copy number variation spanning 60 million years of human and primate evolution. Genome Research, 17(9), 1266-1277.
Freeman, J. L., Perry, G. H., Feuk, L., Redon, R., McCarroll, S. A., Altshuler, D. M., et al. (2006). Copy number variation: New insights in genome diversity. Genome Research, 16(8), 949-961.
Fu, W., Zhang, F., Wang, Y., Gu, X., & Jin, L. (2010). Identification of Copy Number Variation Hotspots in Human Populations. The American Journal of Human Genetics, 87(4), 494-504.
Guirouilh-Barbat, J. e., Huck, S., Bertrand, P., Pirzio, L., Desmaze, C., Sabatier, L., et al. (2004). Impact of the KU80 Pathway on NHEJ-Induced Genome Rearrangements in Mammalian Cells. Molecular Cell, 14(5), 611-623.
Hardwick, R. J., Machado, L. R., Zuccherato, L. W., Antolinos, S., Xue, Y., Shawa, N., et al. (2011). A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Human Mutation, 32(7), 743-750.
Hastings, P. J., Lupski, J. R., Rosenberg, S. M., & Ira, G. (2009). Mechanisms of change in gene copy number. Nature Reviews. Genetics, 10(8), 551-564.
Hawks, J., Wang, E. T., Cochran, G. M., Harpending, H. C., & Moyzis, R. K. (2007). Recent acceleration of human adaptive evolution. Proceedings of the National Academy of Sciences, 104(52), 20753-20758.
Horev, G., Ellegood, J., Lerch, J. P., Son, Y.-E. E., Muthuswamy, L., Vogel, H., et al. (2011). Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proceedings of the National Academy of Sciences, 108(41), 17076-17081.
Lacroix, M. H., Oparina, N. Y., & Mashkova, T. D. (2003). Segmental Duplications in the Human Genome. Molecular Biology, 37(2), 186-193.
Lee, J. A., & Lupski, J. R. (2006). Genomic Rearrangements and Gene Copy-Number Alterations as a Cause of Nervous System Disorders. Neuron, 52, 103-121.
Li, J., Yang, T., Wang, L., Yan, H., Zhang, Y., Guo, Y., et al. (2009). Whole Genome Distribution and Ethnic Differentiation of Copy Number Variation in Caucasian and Asian Populations. PLoS ONE, 4(11), e7958.
Lieber, M. R., Ma, Y., Pannicke, U., & Schwarz, K. (2003). Mechanism and regulation of human non-homologous DNA end-joining. Nature Reviews. Molecular Cell Biology, 4(9), 712-720.
Mandel, A. L., Peyrot des Gachons, C., Plank, K. L., Alarcon, S., & Breslin, P. A. S. (2010). Individual Differences in AMY1 Gene Copy Number, Salivary Amylase Levels, and the Perception of Oral Starch. PLoS ONE, 5(10), e13352.
McLean, C. Y., Reno, P. L., Pollen, A. A., Bassan, A. I., Capellini, T. D., Guenther, C., et al. (2011). Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature, 471(7337), 216-219.
Mills, R. E., Walter, K., Stewart, C., Handsaker, R. E., Chen, K., Alkan, C., et al. (2011). Mapping copy number variation by population-scale genome sequencing. Nature, 470(7332), 59-65.
Navarrete, A., van Schaik, C. P., & Isler, K. (2011). Energetics and the evolution of human brain size. [10.1038/nature10629]. Nature, 480(7375), 91-93.
Novembre, J., Pritchard, J. K., & Coop, G. (2007). Adaptive drool in the gene pool. Nature Genetics, 39(10), 1188-1190.
Perry, G. H. (2008). The evolutionary significance of copy number variation in the human genome. Cytogenetic and Genome Research, 123(1-4), 283-287.
Perry, G. H., Dominy, N. J., Claw, K. G., Lee, A. S., Fiegler, H., Redon, R., et al. (2007). Diet and the evolution of human amylase gene copy number variation. [10.1038/ng2123]. Nat Genet, 39(10), 1256-1260.
Perry, G. H., Yang, F., Marques-Bonet, T., Murphy, C., Fitzgerald, T., Lee, A. S., et al. (2008). Copy number variation and evolution in humans and chimpanzees. Genome Research, 18(11), 1698-1710.
Redon, R., Ishikawa, S., Fitch, K. R., Feuk, L., Perry, G. H., Andrews, T. D., et al. (2006). Global variation in copy number in the human genome. Nature, 444(7118), 444-454.
Richerson, P. J., Boyd, R., & Henrich, J. (2010). Gene-culture coevolution in the age of genomics. Proceedings of the National Academy of Sciences, 107, 8985-8992.
van Driel, M. F. (2011). Re: Human-Specific Loss of Regulatory DNA and the Evolution of Human-Specific Traits. European Urology, 60(5), 1123-1124.
Wang, E. T., Kodama, G., Baldi, P., & Moyzis, R. K. (2006). Global landscape of recent inferred Darwinian selection for Homo sapiens. Proceedings of the National Academy of Sciences of the United States of America, 103(1), 135-140.
Watson, J. (2004). DNA The Secret of Life. London: Arrow Books.
Zhang, F., Gu, W., Hurles, M. E., & Lupski, J. R. (2009). Copy Number Variation in Human Health, Disease, and Evolution. Annual Review of Genomics and Human Genetics, 10(1), 451-481.
Posted in anthropology, biology, evolution, gene-culture coevolution, genetics, primatology
Tags: CNVs, copy number variation, genomics, human evolution, positive selection